
Common Application Areas of bnns
Source:vignettes/articles/common_applications.Rmd
common_applications.Rmd
stopifnot("mlbench not installed" = requireNamespace("mlbench", quietly = TRUE))
stopifnot("rsample not installed" = requireNamespace("rsample", quietly = TRUE))
library(bnns)
library(mlbench)
library(rsample)
set.seed(123)Introduction
This article demonstrates the use of the bnns package on
three datasets from the mlbench package:
-
Regression:
BostonHousingdataset
-
Binary Classification:
PimaIndiansDiabetesdataset
-
Multi-class Classification:
Glassdataset
For each dataset, we: 1. Prepare the data for training and testing.
2. Build a Bayesian Neural Network using the bnns package.
3. Evaluate the model’s predictive performance.
Regression: BostonHousing Dataset
Dataset Description
The BostonHousing dataset contains information on
housing prices in Boston, with features like crime rate, average number
of rooms, and more.
data(BostonHousing)
BH_data <- BostonHousing
# Splitting data into training and testing sets
BH_split <- initial_split(BH_data, prop = 0.8)
BH_train <- training(BH_split)
BH_test <- testing(BH_split)Model Training
model_reg <- bnns(
medv ~ -1 + .,
data = BH_train, L = 2, out_act_fn = 1,
iter = 1e3, warmup = 2e2, chains = 2, cores = 2
)
#> Trying to compile a simple C file
#> Running /opt/R/4.4.2/lib/R/bin/R CMD SHLIB foo.c
#> using C compiler: ‘gcc (Ubuntu 13.3.0-6ubuntu2~24.04) 13.3.0’
#> gcc -I"/opt/R/4.4.2/lib/R/include" -DNDEBUG -I"/home/runner/work/_temp/Library/Rcpp/include/" -I"/home/runner/work/_temp/Library/RcppEigen/include/" -I"/home/runner/work/_temp/Library/RcppEigen/include/unsupported" -I"/home/runner/work/_temp/Library/BH/include" -I"/home/runner/work/_temp/Library/StanHeaders/include/src/" -I"/home/runner/work/_temp/Library/StanHeaders/include/" -I"/home/runner/work/_temp/Library/RcppParallel/include/" -I"/home/runner/work/_temp/Library/rstan/include" -DEIGEN_NO_DEBUG -DBOOST_DISABLE_ASSERTS -DBOOST_PENDING_INTEGER_LOG2_HPP -DSTAN_THREADS -DUSE_STANC3 -DSTRICT_R_HEADERS -DBOOST_PHOENIX_NO_VARIADIC_EXPRESSION -D_HAS_AUTO_PTR_ETC=0 -include '/home/runner/work/_temp/Library/StanHeaders/include/stan/math/prim/fun/Eigen.hpp' -D_REENTRANT -DRCPP_PARALLEL_USE_TBB=1 -I/usr/local/include -fpic -g -O2 -c foo.c -o foo.o
#> In file included from /home/runner/work/_temp/Library/RcppEigen/include/Eigen/Core:19,
#> from /home/runner/work/_temp/Library/RcppEigen/include/Eigen/Dense:1,
#> from /home/runner/work/_temp/Library/StanHeaders/include/stan/math/prim/fun/Eigen.hpp:22,
#> from <command-line>:
#> /home/runner/work/_temp/Library/RcppEigen/include/Eigen/src/Core/util/Macros.h:679:10: fatal error: cmath: No such file or directory
#> 679 | #include <cmath>
#> | ^~~~~~~
#> compilation terminated.
#> make: *** [/opt/R/4.4.2/lib/R/etc/Makeconf:195: foo.o] Error 1
#>
#> SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 0.00037 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 3.7 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#>
#> SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 2).
#> Chain 1: Iteration: 1 / 1000 [ 0%] (Warmup)
#> Chain 2:
#> Chain 2: Gradient evaluation took 0.000458 seconds
#> Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 4.58 seconds.
#> Chain 2: Adjust your expectations accordingly!
#> Chain 2:
#> Chain 2:
#> Chain 2: Iteration: 1 / 1000 [ 0%] (Warmup)
#> Chain 2: Iteration: 100 / 1000 [ 10%] (Warmup)
#> Chain 1: Iteration: 100 / 1000 [ 10%] (Warmup)
#> Chain 2: Iteration: 200 / 1000 [ 20%] (Warmup)
#> Chain 2: Iteration: 201 / 1000 [ 20%] (Sampling)
#> Chain 1: Iteration: 200 / 1000 [ 20%] (Warmup)
#> Chain 1: Iteration: 201 / 1000 [ 20%] (Sampling)
#> Chain 2: Iteration: 300 / 1000 [ 30%] (Sampling)
#> Chain 2: Iteration: 400 / 1000 [ 40%] (Sampling)
#> Chain 1: Iteration: 300 / 1000 [ 30%] (Sampling)
#> Chain 2: Iteration: 500 / 1000 [ 50%] (Sampling)
#> Chain 1: Iteration: 400 / 1000 [ 40%] (Sampling)
#> Chain 2: Iteration: 600 / 1000 [ 60%] (Sampling)
#> Chain 2: Iteration: 700 / 1000 [ 70%] (Sampling)
#> Chain 1: Iteration: 500 / 1000 [ 50%] (Sampling)
#> Chain 2: Iteration: 800 / 1000 [ 80%] (Sampling)
#> Chain 2: Iteration: 900 / 1000 [ 90%] (Sampling)
#> Chain 1: Iteration: 600 / 1000 [ 60%] (Sampling)
#> Chain 2: Iteration: 1000 / 1000 [100%] (Sampling)
#> Chain 2:
#> Chain 2: Elapsed Time: 15.616 seconds (Warm-up)
#> Chain 2: 48.368 seconds (Sampling)
#> Chain 2: 63.984 seconds (Total)
#> Chain 2:
#> Chain 1: Iteration: 700 / 1000 [ 70%] (Sampling)
#> Chain 1: Iteration: 800 / 1000 [ 80%] (Sampling)
#> Chain 1: Iteration: 900 / 1000 [ 90%] (Sampling)
#> Chain 1: Iteration: 1000 / 1000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 18.408 seconds (Warm-up)
#> Chain 1: 81.008 seconds (Sampling)
#> Chain 1: 99.416 seconds (Total)
#> Chain 1:Model Evaluation
BH_pred <- predict(model_reg, newdata = BH_test)
measure_cont(BH_test$medv, BH_pred)
#> $rmse
#> [1] 8.297719
#>
#> $mae
#> [1] 5.822318Binary Classification: PimaIndiansDiabetes Dataset
Dataset Description
The PimaIndiansDiabetes dataset contains features
related to health status for predicting the presence of diabetes.
Model Training
model_bin <- bnns(
diabetes ~ -1 + .,
data = PID_train, L = 2,
out_act_fn = 2, iter = 1e3, warmup = 2e2, chains = 2, cores = 2
)
#> Trying to compile a simple C file
#> Running /opt/R/4.4.2/lib/R/bin/R CMD SHLIB foo.c
#> using C compiler: ‘gcc (Ubuntu 13.3.0-6ubuntu2~24.04) 13.3.0’
#> gcc -I"/opt/R/4.4.2/lib/R/include" -DNDEBUG -I"/home/runner/work/_temp/Library/Rcpp/include/" -I"/home/runner/work/_temp/Library/RcppEigen/include/" -I"/home/runner/work/_temp/Library/RcppEigen/include/unsupported" -I"/home/runner/work/_temp/Library/BH/include" -I"/home/runner/work/_temp/Library/StanHeaders/include/src/" -I"/home/runner/work/_temp/Library/StanHeaders/include/" -I"/home/runner/work/_temp/Library/RcppParallel/include/" -I"/home/runner/work/_temp/Library/rstan/include" -DEIGEN_NO_DEBUG -DBOOST_DISABLE_ASSERTS -DBOOST_PENDING_INTEGER_LOG2_HPP -DSTAN_THREADS -DUSE_STANC3 -DSTRICT_R_HEADERS -DBOOST_PHOENIX_NO_VARIADIC_EXPRESSION -D_HAS_AUTO_PTR_ETC=0 -include '/home/runner/work/_temp/Library/StanHeaders/include/stan/math/prim/fun/Eigen.hpp' -D_REENTRANT -DRCPP_PARALLEL_USE_TBB=1 -I/usr/local/include -fpic -g -O2 -c foo.c -o foo.o
#> In file included from /home/runner/work/_temp/Library/RcppEigen/include/Eigen/Core:19,
#> from /home/runner/work/_temp/Library/RcppEigen/include/Eigen/Dense:1,
#> from /home/runner/work/_temp/Library/StanHeaders/include/stan/math/prim/fun/Eigen.hpp:22,
#> from <command-line>:
#> /home/runner/work/_temp/Library/RcppEigen/include/Eigen/src/Core/util/Macros.h:679:10: fatal error: cmath: No such file or directory
#> 679 | #include <cmath>
#> | ^~~~~~~
#> compilation terminated.
#> make: *** [/opt/R/4.4.2/lib/R/etc/Makeconf:195: foo.o] Error 1
#>
#> SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 0.000439 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 4.39 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#>
#> SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 2).
#> Chain 1: Iteration: 1 / 1000 [ 0%] (Warmup)
#> Chain 2:
#> Chain 2: Gradient evaluation took 0.000422 seconds
#> Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 4.22 seconds.
#> Chain 2: Adjust your expectations accordingly!
#> Chain 2:
#> Chain 2:
#> Chain 2: Iteration: 1 / 1000 [ 0%] (Warmup)
#> Chain 1: Iteration: 100 / 1000 [ 10%] (Warmup)
#> Chain 2: Iteration: 100 / 1000 [ 10%] (Warmup)
#> Chain 1: Iteration: 200 / 1000 [ 20%] (Warmup)
#> Chain 1: Iteration: 201 / 1000 [ 20%] (Sampling)
#> Chain 2: Iteration: 200 / 1000 [ 20%] (Warmup)
#> Chain 2: Iteration: 201 / 1000 [ 20%] (Sampling)
#> Chain 2: Iteration: 300 / 1000 [ 30%] (Sampling)
#> Chain 1: Iteration: 300 / 1000 [ 30%] (Sampling)
#> Chain 2: Iteration: 400 / 1000 [ 40%] (Sampling)
#> Chain 1: Iteration: 400 / 1000 [ 40%] (Sampling)
#> Chain 2: Iteration: 500 / 1000 [ 50%] (Sampling)
#> Chain 1: Iteration: 500 / 1000 [ 50%] (Sampling)
#> Chain 2: Iteration: 600 / 1000 [ 60%] (Sampling)
#> Chain 2: Iteration: 700 / 1000 [ 70%] (Sampling)
#> Chain 1: Iteration: 600 / 1000 [ 60%] (Sampling)
#> Chain 2: Iteration: 800 / 1000 [ 80%] (Sampling)
#> Chain 1: Iteration: 700 / 1000 [ 70%] (Sampling)
#> Chain 2: Iteration: 900 / 1000 [ 90%] (Sampling)
#> Chain 1: Iteration: 800 / 1000 [ 80%] (Sampling)
#> Chain 2: Iteration: 1000 / 1000 [100%] (Sampling)
#> Chain 2:
#> Chain 2: Elapsed Time: 24.285 seconds (Warm-up)
#> Chain 2: 63.093 seconds (Sampling)
#> Chain 2: 87.378 seconds (Total)
#> Chain 2:
#> Chain 1: Iteration: 900 / 1000 [ 90%] (Sampling)
#> Chain 1: Iteration: 1000 / 1000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 22.383 seconds (Warm-up)
#> Chain 1: 82.263 seconds (Sampling)
#> Chain 1: 104.646 seconds (Total)
#> Chain 1:Model Evaluation
PID_pred <- predict(model_bin, newdata = PID_test)
PID_measure <- measure_bin(PID_test$diabetes, PID_pred)
#> Setting levels: control = 0, case = 1
#> Setting direction: controls < cases
PID_measure
#> $conf_mat
#> pred_label
#> obs 0 1
#> 0 82 18
#> 1 29 25
#>
#> $accuracy
#> [1] 0.6948052
#>
#> $ROC
#>
#> Call:
#> roc.default(response = obs, predictor = pred)
#>
#> Data: pred in 100 controls (obs 0) < 54 cases (obs 1).
#> Area under the curve: 0.7296
#>
#> $AUC
#> [1] 0.7296296
plot(PID_measure$ROC)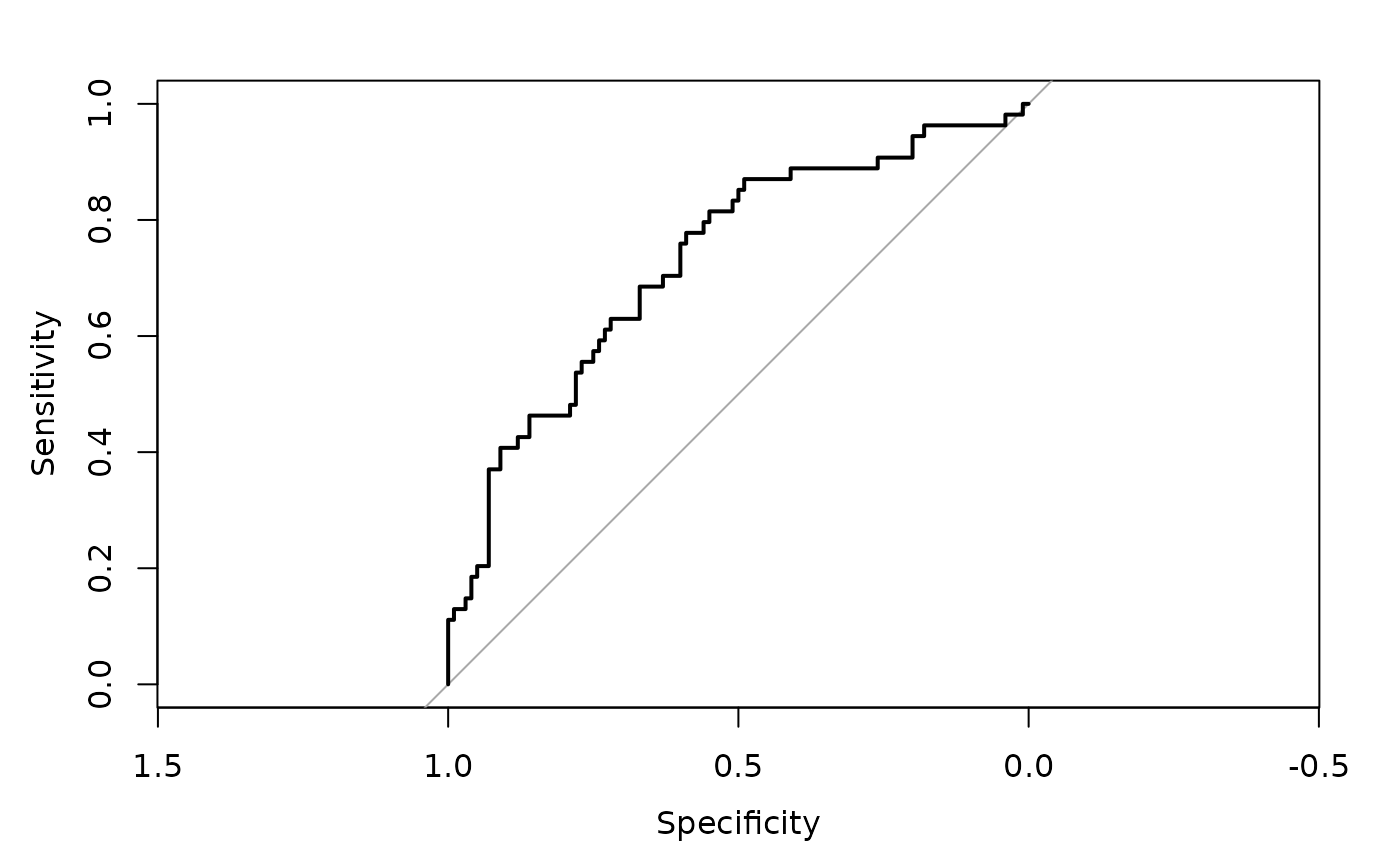
Multi-class Classification: Glass Dataset
Dataset Description
The Glass dataset contains features to classify glass
types.
data(Glass)
Glass_data <- Glass
# Splitting data into training and testing sets
Glass_split <- initial_split(Glass_data, prop = 0.8, strata = "Type")
Glass_train <- training(Glass_split)
Glass_test <- testing(Glass_split)Model Training
model_multi <- bnns(
Type ~ -1 + .,
data = Glass_train, L = 2,
out_act_fn = 3, iter = 1e3, warmup = 2e2, chains = 2, cores = 2
)
#> Trying to compile a simple C file
#> Running /opt/R/4.4.2/lib/R/bin/R CMD SHLIB foo.c
#> using C compiler: ‘gcc (Ubuntu 13.3.0-6ubuntu2~24.04) 13.3.0’
#> gcc -I"/opt/R/4.4.2/lib/R/include" -DNDEBUG -I"/home/runner/work/_temp/Library/Rcpp/include/" -I"/home/runner/work/_temp/Library/RcppEigen/include/" -I"/home/runner/work/_temp/Library/RcppEigen/include/unsupported" -I"/home/runner/work/_temp/Library/BH/include" -I"/home/runner/work/_temp/Library/StanHeaders/include/src/" -I"/home/runner/work/_temp/Library/StanHeaders/include/" -I"/home/runner/work/_temp/Library/RcppParallel/include/" -I"/home/runner/work/_temp/Library/rstan/include" -DEIGEN_NO_DEBUG -DBOOST_DISABLE_ASSERTS -DBOOST_PENDING_INTEGER_LOG2_HPP -DSTAN_THREADS -DUSE_STANC3 -DSTRICT_R_HEADERS -DBOOST_PHOENIX_NO_VARIADIC_EXPRESSION -D_HAS_AUTO_PTR_ETC=0 -include '/home/runner/work/_temp/Library/StanHeaders/include/stan/math/prim/fun/Eigen.hpp' -D_REENTRANT -DRCPP_PARALLEL_USE_TBB=1 -I/usr/local/include -fpic -g -O2 -c foo.c -o foo.o
#> In file included from /home/runner/work/_temp/Library/RcppEigen/include/Eigen/Core:19,
#> from /home/runner/work/_temp/Library/RcppEigen/include/Eigen/Dense:1,
#> from /home/runner/work/_temp/Library/StanHeaders/include/stan/math/prim/fun/Eigen.hpp:22,
#> from <command-line>:
#> /home/runner/work/_temp/Library/RcppEigen/include/Eigen/src/Core/util/Macros.h:679:10: fatal error: cmath: No such file or directory
#> 679 | #include <cmath>
#> | ^~~~~~~
#> compilation terminated.
#> make: *** [/opt/R/4.4.2/lib/R/etc/Makeconf:195: foo.o] Error 1
#>
#> SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 0.000343 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 3.43 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 1000 [ 0%] (Warmup)
#>
#> SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 2).
#> Chain 2:
#> Chain 2: Gradient evaluation took 0.00032 seconds
#> Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 3.2 seconds.
#> Chain 2: Adjust your expectations accordingly!
#> Chain 2:
#> Chain 2:
#> Chain 2: Iteration: 1 / 1000 [ 0%] (Warmup)
#> Chain 1: Iteration: 100 / 1000 [ 10%] (Warmup)
#> Chain 2: Iteration: 100 / 1000 [ 10%] (Warmup)
#> Chain 1: Iteration: 200 / 1000 [ 20%] (Warmup)
#> Chain 1: Iteration: 201 / 1000 [ 20%] (Sampling)
#> Chain 2: Iteration: 200 / 1000 [ 20%] (Warmup)
#> Chain 2: Iteration: 201 / 1000 [ 20%] (Sampling)
#> Chain 1: Iteration: 300 / 1000 [ 30%] (Sampling)
#> Chain 2: Iteration: 300 / 1000 [ 30%] (Sampling)
#> Chain 1: Iteration: 400 / 1000 [ 40%] (Sampling)
#> Chain 1: Iteration: 500 / 1000 [ 50%] (Sampling)
#> Chain 2: Iteration: 400 / 1000 [ 40%] (Sampling)
#> Chain 1: Iteration: 600 / 1000 [ 60%] (Sampling)
#> Chain 2: Iteration: 500 / 1000 [ 50%] (Sampling)
#> Chain 1: Iteration: 700 / 1000 [ 70%] (Sampling)
#> Chain 1: Iteration: 800 / 1000 [ 80%] (Sampling)
#> Chain 2: Iteration: 600 / 1000 [ 60%] (Sampling)
#> Chain 1: Iteration: 900 / 1000 [ 90%] (Sampling)
#> Chain 1: Iteration: 1000 / 1000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 16.97 seconds (Warm-up)
#> Chain 1: 53.068 seconds (Sampling)
#> Chain 1: 70.038 seconds (Total)
#> Chain 1:
#> Chain 2: Iteration: 700 / 1000 [ 70%] (Sampling)
#> Chain 2: Iteration: 800 / 1000 [ 80%] (Sampling)
#> Chain 2: Iteration: 900 / 1000 [ 90%] (Sampling)
#> Chain 2: Iteration: 1000 / 1000 [100%] (Sampling)
#> Chain 2:
#> Chain 2: Elapsed Time: 18.752 seconds (Warm-up)
#> Chain 2: 84.929 seconds (Sampling)
#> Chain 2: 103.681 seconds (Total)
#> Chain 2:Model Evaluation
Glass_pred <- predict(model_multi, newdata = Glass_test)
measure_cat(Glass_test$Type, Glass_pred)
#> $log_loss
#> [1] 1.353789
#>
#> $ROC
#>
#> Call:
#> multiclass.roc.default(response = obs, predictor = `colnames<-`(data.frame(pred), levels(obs)))
#>
#> Data: multivariate predictor `colnames<-`(data.frame(pred), levels(obs)) with 6 levels of obs: 1, 2, 3, 5, 6, 7.
#> Multi-class area under the curve: 0.7095
#>
#> $AUC
#> [1] 0.7094742